
The Genome Stability and Editing Laboratory

RESEARCH
Susumu Tonegawa, 1987 Nobel Prize in Medicine: "Creativity is made of 3 simple concepts: 1) You have to be very curious about something; 2) You have to have unfailing urge to address the question you have; 3) Try to combine knowledge in at least two different fields where people don't necessarily interact."
Our laboratory is studying the mechanisms that promote DNA Repair and Genome Editing.
Our laboratory is at the forefront of unlocking the secrets of DNA repair and genome editing, using state-of-the-art technologies to manipulate the very building blocks of life. Through high-throughput genetic screens, biochemical assays, and cell biology experiments, we are unraveling the complex mechanisms that protect our genome and ensure its stability.
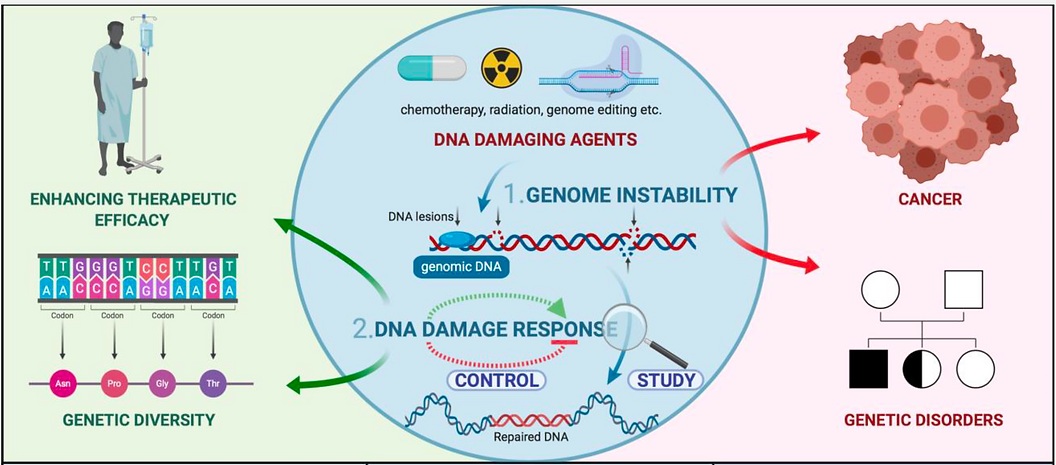
Our mission is nothing less than to unlock the secrets of the cellular DNA damage response, and at developing genome editing technologies, two critical areas of research that have far-reaching implications for the future of medicine and biotechnology.
GENOME EDITING - MAKING THE WAY FOR PRECISION THERAPEUTICS.
DNA is the cornerstone of all living organisms, carrying invaluable genetic information. However, genetic variants present in the genome can lead to devastating diseases. That's where genome editing comes in - a revolutionary technology that holds the power to cure or even prevent these life-threatening conditions by eliminating harmful genetic variants.
By triggering specific cellular DNA repair systems, genome editing tools such as engineered nucleases and CRISPR-associated nucleases have unlocked the potential for targeted and precise genetic modifications. Modern genome editing agents like programmable base editors, prime-editors, and integrases are able to introduce precise base modifications and insertions/deletions, providing a path for accurate editing with increased efficiency, safety, and predictability.
The ability to manipulate the cellular DNA repair mechanisms not only opens up new avenues for precision therapeutics but also deepens our understanding of fundamental DNA repair mechanisms. This knowledge will help us model and study pathogenic variants with greater precision and accuracy.
Our recent work in this area is available here.
Modern CRISPR-based genome editing technologies.
Several site-specific DNA lesions, including double-strand breaks, nicks, deaminated bases and flaps, are induced by modern CRISPR-based genome editing technologies that are detected and repaired by the DDR resulting in editing.
a) CRISPR-Cas9. Cas9 initiates genome editing by introducing a site-specific double-strand break.
b) Base editing. Nickase Cas9 is fused to deaminases for targeted base deamination.
c) Prime editing. Nickase Cas9 is fused to an engineered reverse transcriptase that directly copies the information contained in the guide RNA.
UNLOCKING THE POWER OF DNA REPAIR IN THE FIGHT AGAINST CANCER
DNA damage response pathways being targeted in the clinic.
DNA repair pathways (MMR, SSB//BER, HR, NHEJ) are activated by various DNA lesions (mismatch, single-strand or double-strand breaks). DNA repair factors are targeted by small molecule inhibitors to induce genomic instability in a range of human cancers.
Unlocking the mysteries of DNA repair is an imperative mission in the battle against cancer, a ruthless disease that has ravaged countless lives. Genomic instability, a hallmark of cancer, represents a daunting challenge that demands a sophisticated solution. Fortunately, cells have evolved intricate and refined mechanisms known as the DNA Damage Response (DDR), which can detect and repair DNA damage, thereby safeguarding the integrity of the genome.
The DDR plays a pivotal role in suppressing tumorigenesis, as well as regulating the response to cancer treatments, by creating exploitable weaknesses in tumor cells. Therefore, a comprehensive understanding of the DDR's complex workings under different conditions is vital for developing targeted therapeutic strategies to combat cancer.
Our laboratory is dedicated to leveraging the power of the DDR to gain a deeper understanding of the underlying mechanisms driving cancer progression and improving cancer therapy. With the help of cutting-edge genome editing technologies, we can now scrutinize the molecular mechanisms governing genome stability at an unprecedented level of precision. These advancements offer a wealth of opportunities to develop new and innovative approaches to fighting cancer by enhancing DNA damage-based treatments and accelerating drug discovery.
To learn more about our work, check out our recent publications here,

